A leading group of researchers have developed a new way to study bowel cancer samples using digital pathology images. This means the subtype of bowel cancer a patient has can be more easily identified to better understand how bowel cancer will grow and respond to treatment.
A leading group of researchers have developed a new way to study bowel cancer samples using digital pathology images. This means the subtype of bowel cancer a patient has can be more easily identified to better understand how bowel cancer will grow and respond to treatment. As research develops, this means patients could be better matched to the best treatment for them.
The study - part of the S:CORT (Stratification in COloRecTal Cancer) consortium - is looking for better ways to make treatment decisions and predict how well these treatments will work for patients with bowel cancer
Why is this sort of research needed?
Our landmark critical research gaps initiative revealed that one of the key bowel cancer research gaps centres around pathology – an area that underpins every aspect of patient care and plays a major role in research. Finding ways to better predict how an individual’s bowel cancer is likely to grow and respond to treatment is critically important.
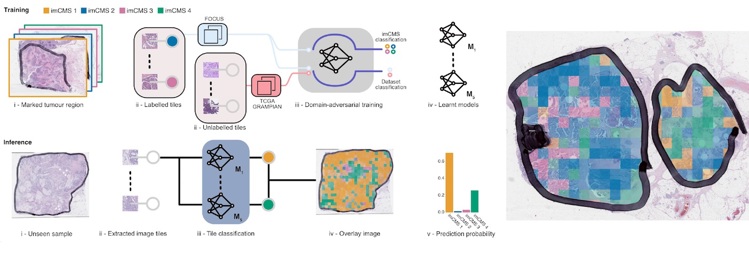
In recent years, researchers have uncovered more about the complex biological details that make up individual bowel cancers – and rather than one disease, it’s now thought that there are four distinct subtypes of the disease – referred to as ‘consensus molecular classification’ (CMS).
Expanding knowledge about the complex parts that make up cancer cells has already enabled the development of cutting edge treatments – so-called ‘targeted therapies’. But more research is needed to fully use these learnings to transform diagnosis, treatment and care for more bowel cancer patients.
The evolution of digital pathology
Alongside progress in our understanding of the make-up of bowel cancer tumours, the way pathologists can actually see these differences has drastically improved too. Much like the way google maps has revolutionised the way we get around compared to a simple paper map, this approach adds computing to use of the microscope. Over the past hundred years, the microscope has been the biggest asset to a pathologist’s toolkit, but using digital images which computers can read has created great leaps forward.
Making sense of these detailed images takes a lot of skill and time, so specialised technology holds real benefit and reduces chance of mistakes. Previously pathologists have been able to diagnose cancer but not identify the subtypes. This research makes it possible to idenitfy the molecular subtype from a routine image of the cancer, providing a better idea of how an individual’s cancer might grow and respond to treatment.
Unlocking the important information encoded within these highly detailed images on a larger scale paves the way for richer, bigger-picture patterns about the different bowel cancers subtypes.
What does this new research add?
"Our approach makes it possible to assist pathologists in a way that has not been possible before"
Using a sophisticated technique termed ‘machine learning’ and alongside their teams spanning several countries, lead researchers Professor Jens Rittscher (Department of Engineering Science, Oxford) and Professor Viktor Koelzer (University of Zurich) trained a computer model to analyse digital images of bowel cancer samples.
The samples they used are collected routinely in hospitals already, but this was the first time this type of computer technology has used digital images to group samples in this way. It means computer analysis of pathology images can be used to understand how a bowel cancer tumour will behave in an individual patient.
Bridging the gap between the complex differences that make up individual bowel cancers and making more tailored decisions about treatment and care is the long term goal – and the study’s researchers hope their work will help accelerate progress towards this.
Professor Rittscher says, "Our ability to identify histology patterns that are biologically relevant and were previously not accessible to human interpretation make this work very exciting. Our approach makes it possible to assist pathologists in a way that has not been possible before. This way we have effectively linked an established molecular disease stratification with traditional pathology. We have already received requests to apply imCMS to retrospective cohorts for which no genetic information is available."
What next?
Applying this technology to large clinical trial datasets will be the first step to see how this can help advance understanding in this area. But making this available routinely in hospitals requires investment in digital pathology services. This revolution is beginning already and is crucial to translating these important learnings into meaningful change to the way bowel cancer is diagnosed and treated in the clinic. Although steps need to be taken to make sure pathology services are ready, hospitals wouldn’t necessarily need this advanced technology on site – in theory, the images could be sent anywhere else in the world for analysis. Further research will be needed to work out which patients might benefit most from this sort of testing and when.
Related paper: Image-based consensus molecular subtype classification (imCMS) of colorectal cancer using deep learning
Content reproduced by kind permission of Bowel Cancer UK
Could fusion solve our clean energy problem?
Oxford University Spinout

